Help
Metadata Portal (NeuroMorpho.Org)
In order to maximize impact and discovery in research, shared data should be annotated with appropriate metadata (information that provides supplementary knowledge about the associated datasets). This online metadata portal for NeuroMorpho.Org relies on state-of-the-art web technologies and is designed to incorporate novel machine learning algorithms for promoting and facilitating the systematic acquisition of descriptive information for neuronal reconstructions. This project is fully open-source and the development may also be extended to other research projects that require knowledge management. Find more about it here.
What is NeuroMorpho.Org?
NeuroMorpho.Org is a centrally curated inventory of digitally reconstructed neurons and glia. It contains contributions from hundreds of laboratories worldwide and is continuously updated as new morphological reconstructions are collected, published, and shared. The goal of NeuroMorpho.Org is to provide free access to all available neuronal reconstruction data in the neuroscience scientific community.
Step by Step procedure to share neuronal reconstructions with NeuroMorpho.Org
This tutorial walks you through basic components of Metadata Portal and shows how to create a dataset, upload recostructions and request for your data to be shared with sientific comunity on NeuroMorpho.Org. Click on the pictures for a bigger image.
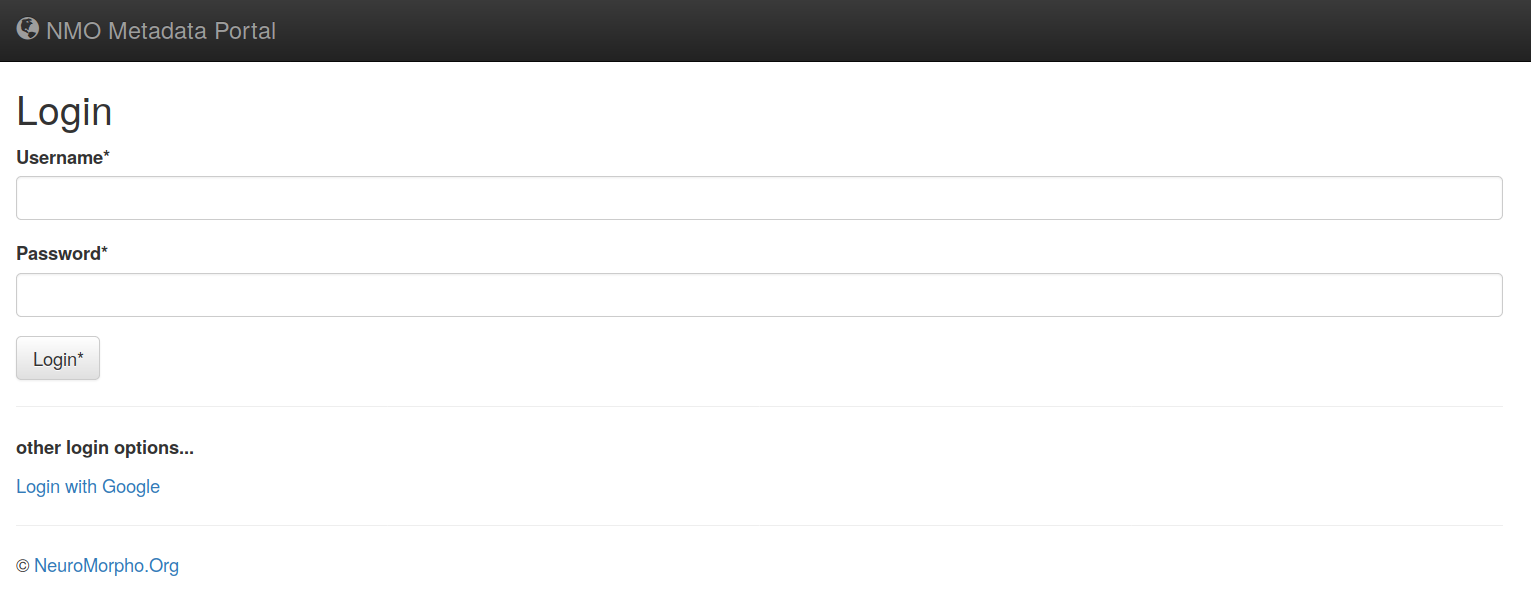
1- Loging credentials for Metadata Portal (http://cng-nmo-meta.orc.gmu.edu/login/)
In order to protect and save the portal from malicious activities, one should log into the system via username and password or using Gmail social login.
You can use our public credentials (username: nmo-author, pass: neuromorpho) [all lower-case] to login to the portal, alternatively, you can use login using you Google account (Login with Google). The latter way is more secure and on the top of everything else, using this method your data is private and won't be visible by anyone else until the final NMO curation process is done and it is shared on NeuroMorpho.Org
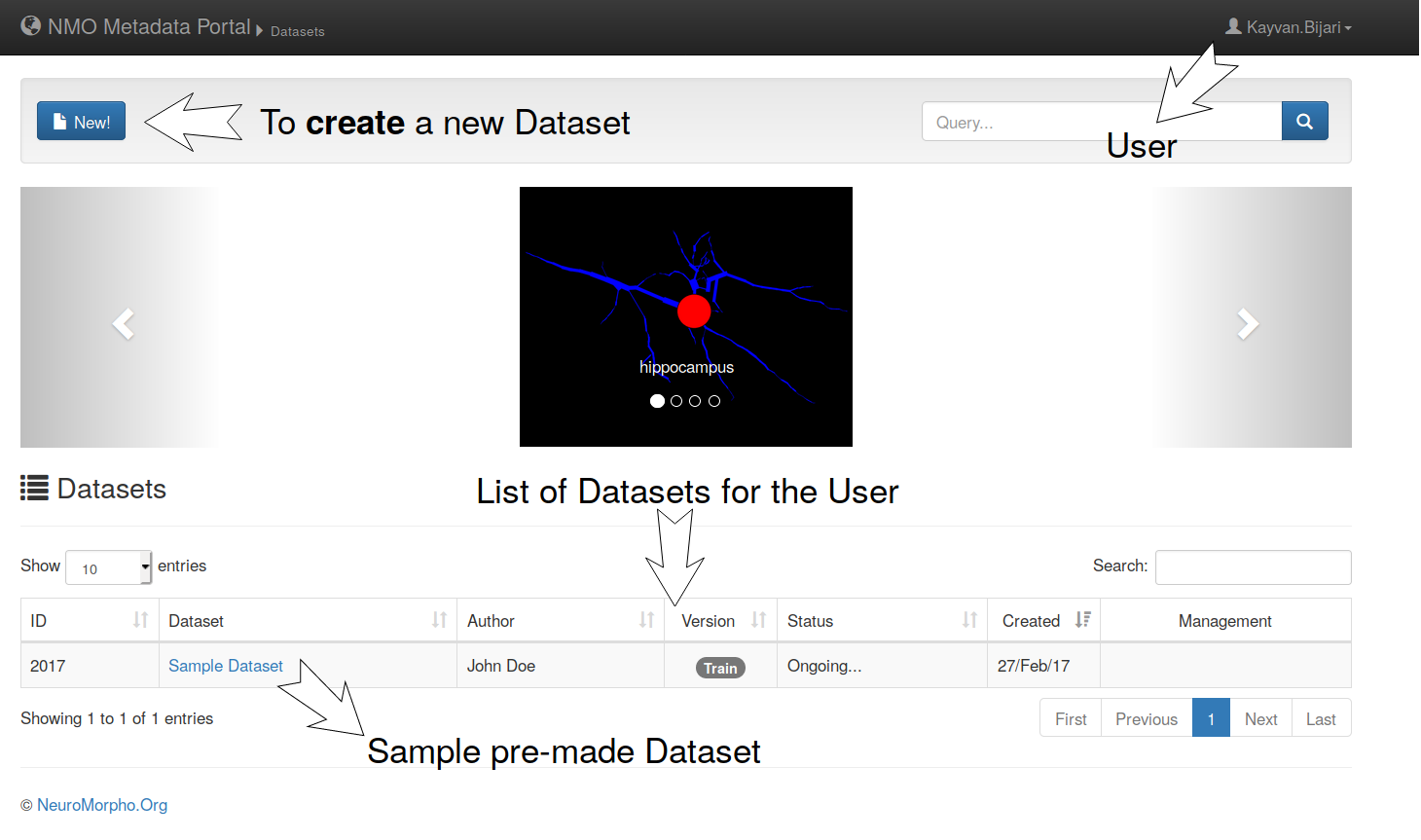
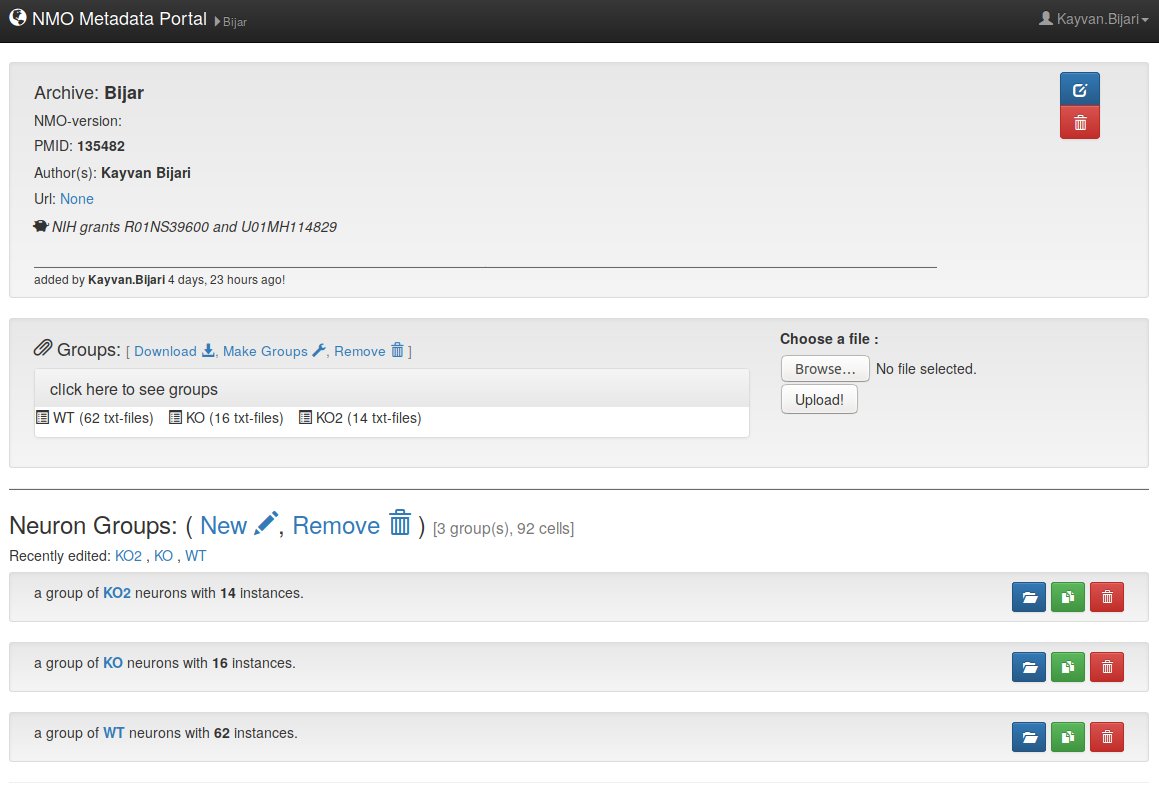
2- Portal overview
– Once you login, you will see the following features on the portal.
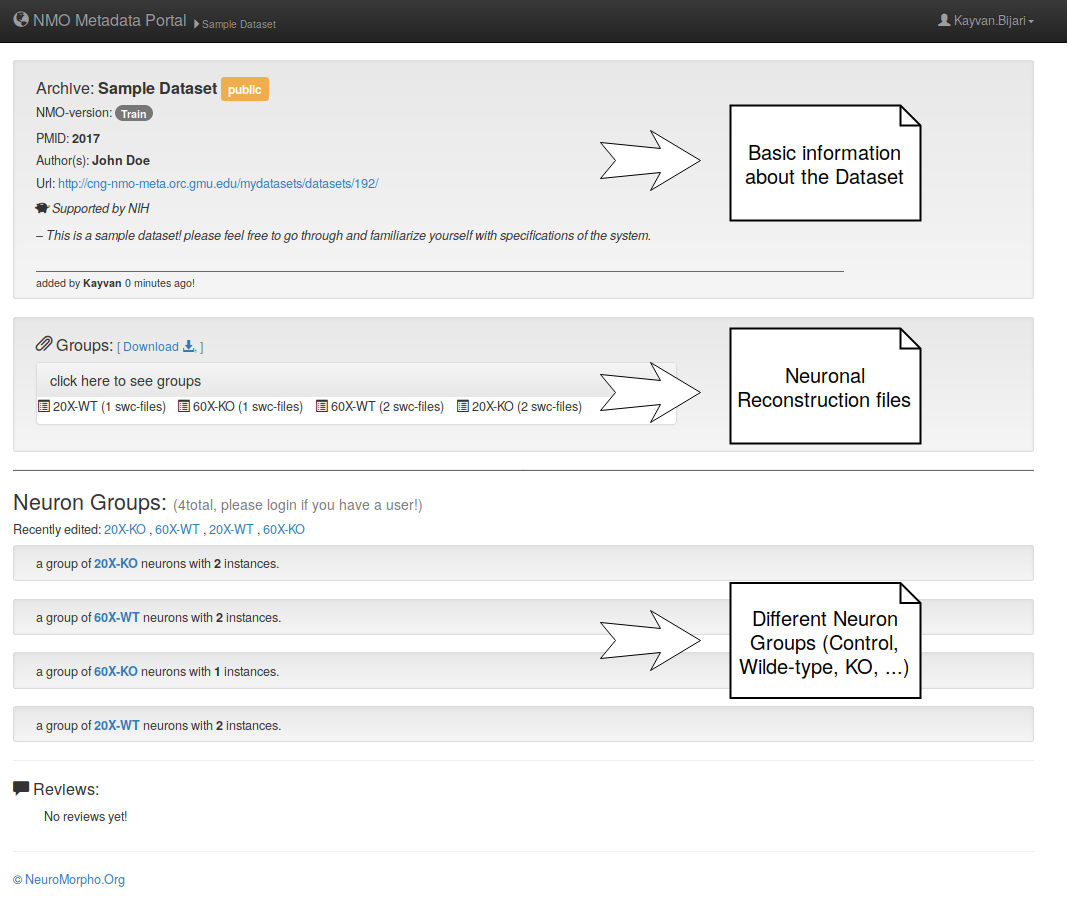
– Here is what a sample dataset entails.
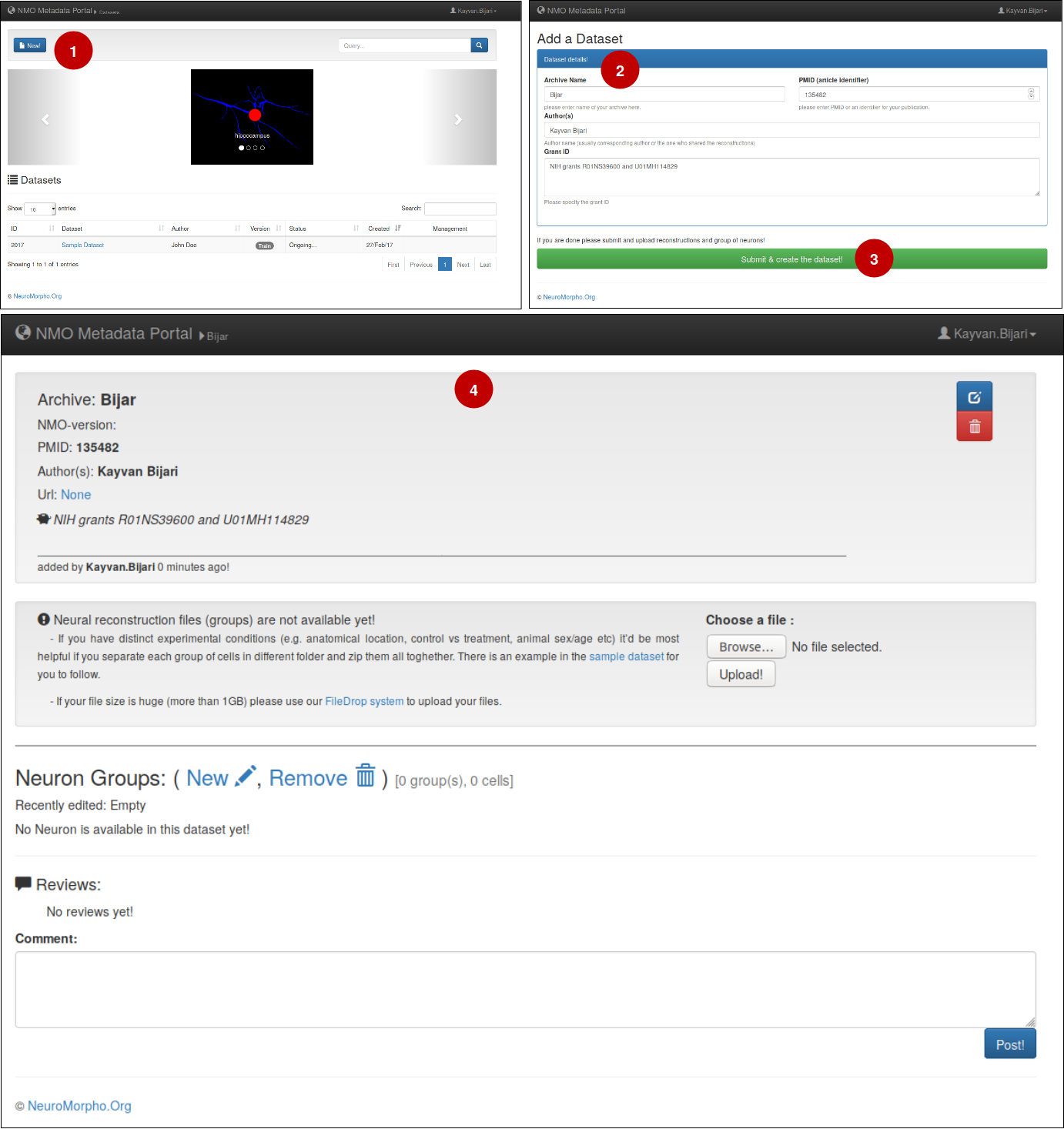
3- Create a Dataset
– To create a new dataset of your own (1) click on the 'New!' in the main view of the portal, (2) then, in the newly opened window insert information related to your publication such as PMID, Author's name, and grand ID. Finally (3) click on 'Submit & create the dataset'. (4) As a result, you will your dataset with basic information that you have entered, and it is ready for the next step (uploading reconstruction files and adding neuron groups).
– At any point, the basic information of the datasets are changeable, to do so, click on the edit icon above the trash icon, editing window pops up and you can simply edit the inserted information.
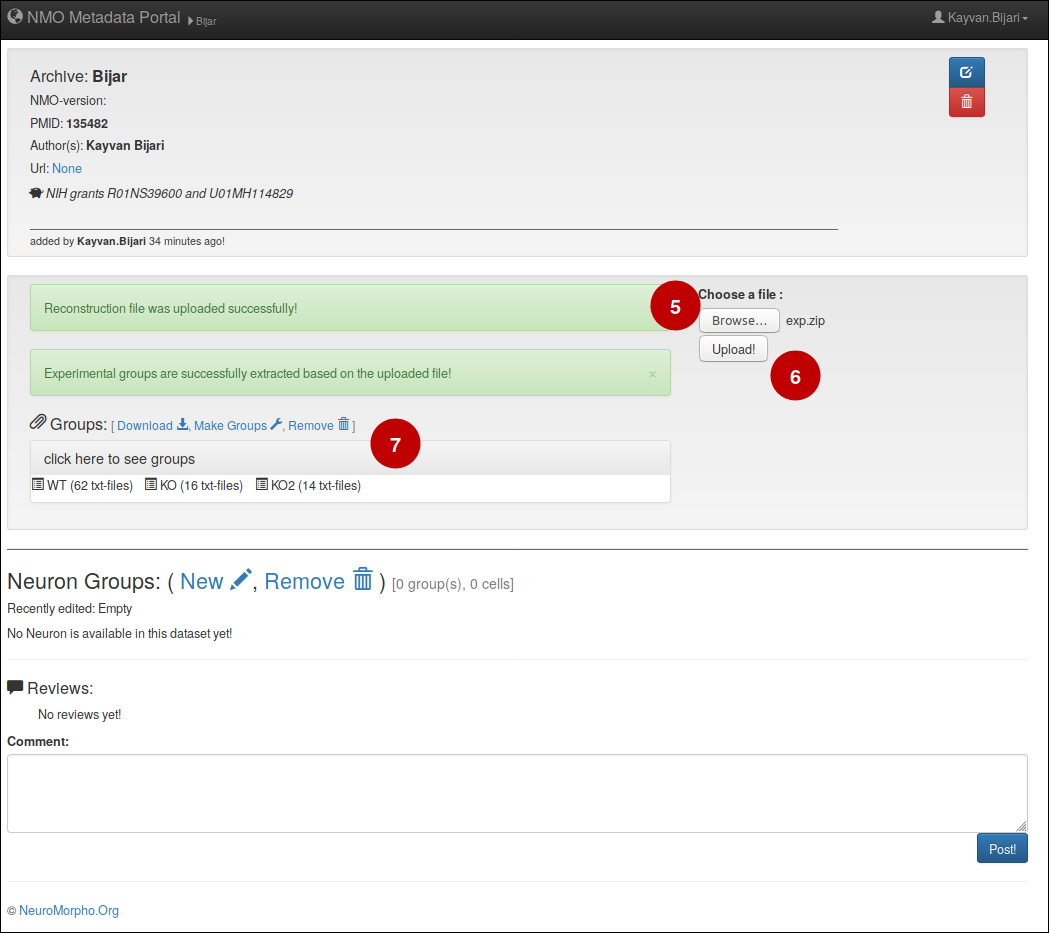
4- Upload reconstruction files
– Please zip your reconstruction files and give a relatively short name to the file (Data.zip, Archive.zip, Reconstructions.zip).
– To upload the reconstruction files, (5) simply click on brows button and select them from your computer, (6) afterwards click on upload button to start the uploading process. (7) You will then see the uploaded files and different experimental groups.
– If you have distinct experimental conditions (e.g. anatomical location, control vs treatment, animal sex/age etc) it’d be most helpful if you separate each group of cells in a different folder and zip them all together.
– If your file size is huge (more than 1GB) please use our FileDrop system to upload your files.
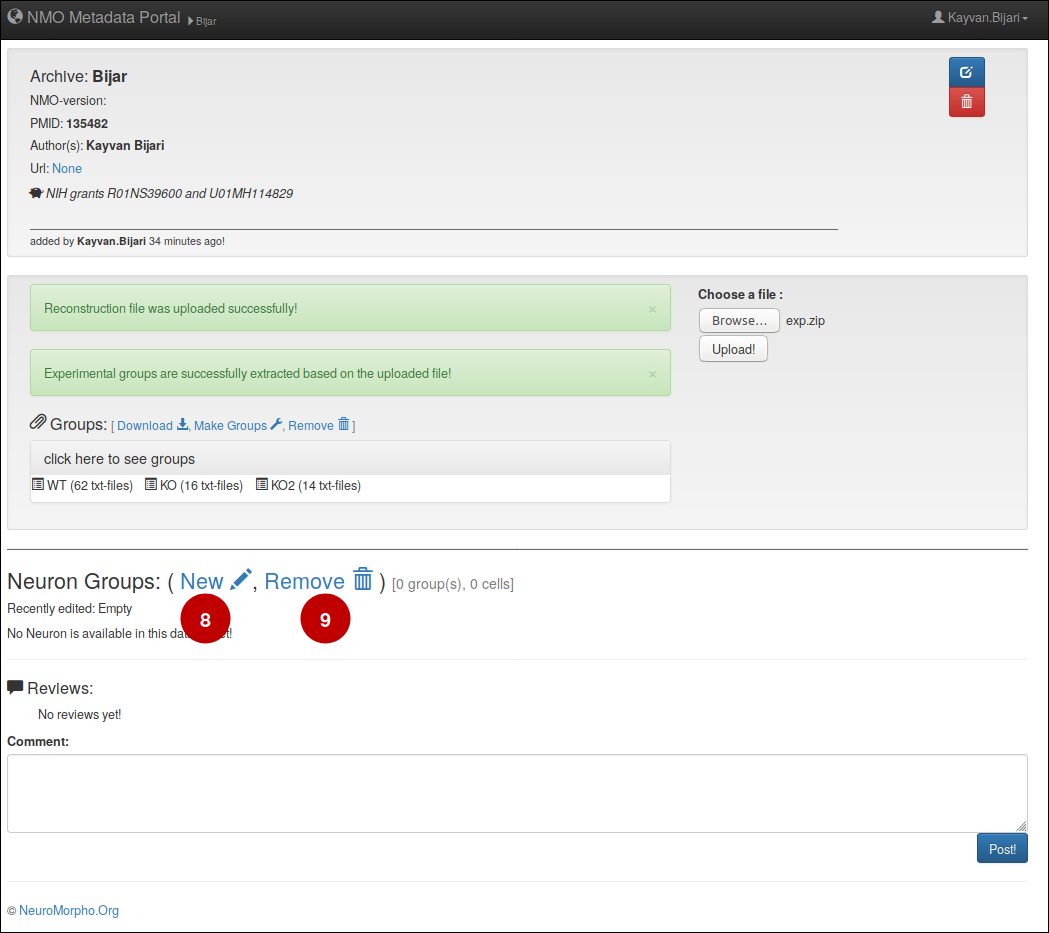
5- Make Groups
– To make a new group, (8) click on 'New', then a new form window pops up and asks you the details of your experimental method. Please note that (9) 'Remove' button will remove all the available groups in the dataset.
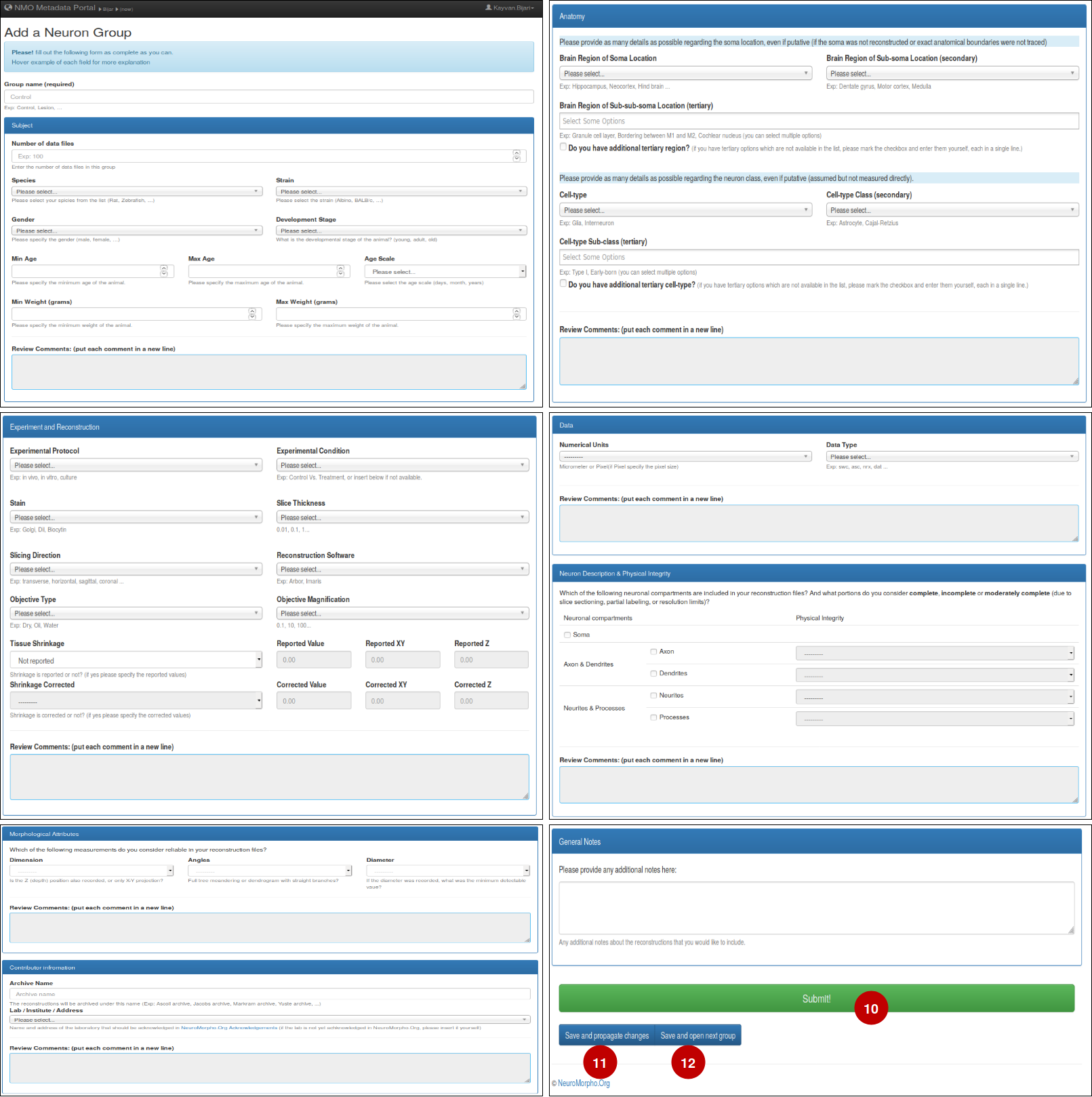
– This is the form which asks for the details of your experiment. Fill out the entries as complete as you can and pay attention to the help messages provided by the portal. Once you're done with all the fields (10) click and submit the group.
– If you have more than one group in the portal, (11) will propagate all changes to all other groups and (12) saves the changes and opens up the next group for you.
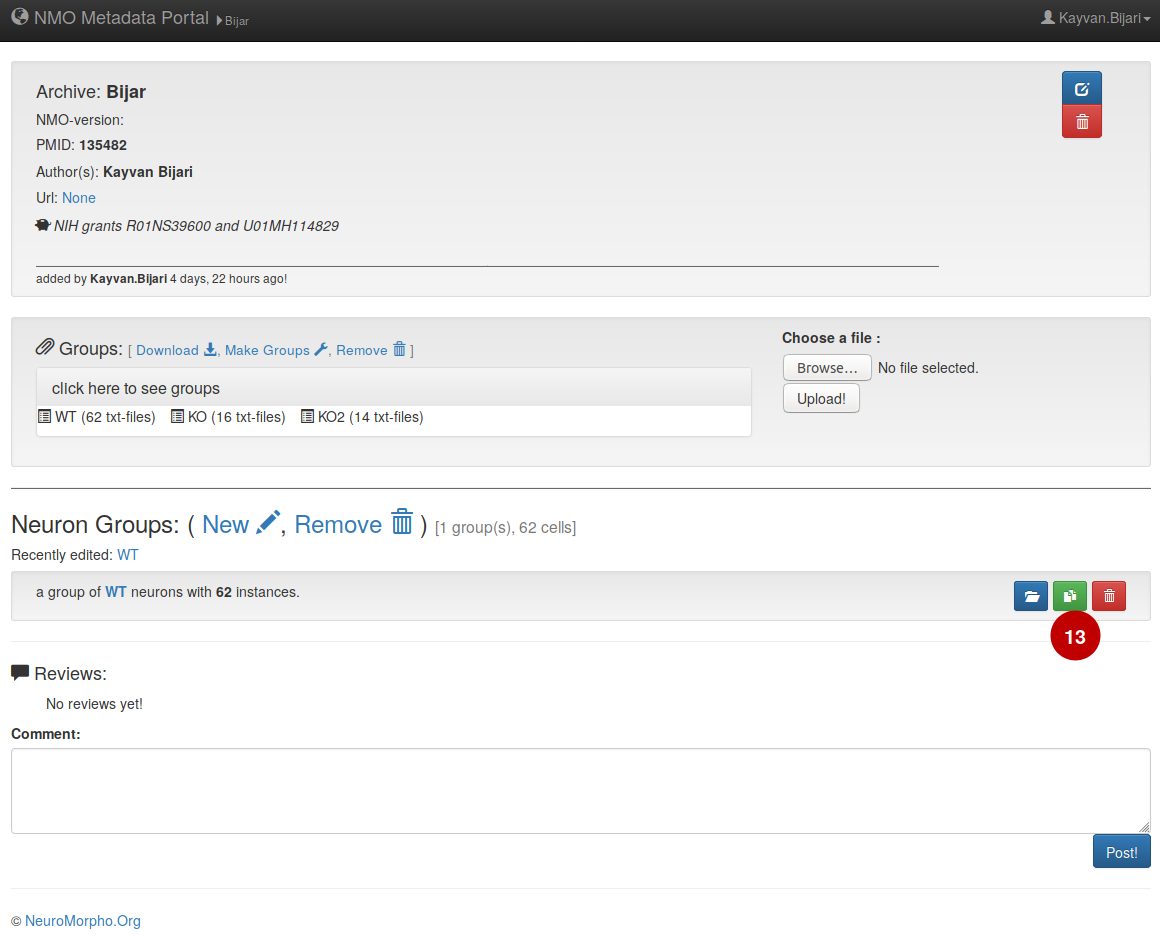
– Once the group is submitted, it will be added under group lists in the portal. If any change needs to be done in the group specification, (13) simply open and edit the group and then submit it when the desired changes are applied.
– In order to add new groups there is two options, one is to add a new group and the procedure is exact same as previous steps. (13) The other option is to duplicate one of the previous groups and then edit/modify the specification of the duplicated group, since a majority of the experimental conditions are the same across different groups duplication of the groups would be a better choice.
– Here is how the dataset should look like finally when reconstructions and all the groups are added.
 drosophila (peripheral nervous system)
drosophila (peripheral nervous system)